Contracted Gaussian type orbitals
Minimal basis sets
Single Gaussian functions as
described by equ. (5) are not well suited to describe the spatial extend and
nodal characteristics of atomic orbitals. To solve this problem, basis
functions are described as a sum (a "contraction") of several Gaussian functions
(primitives):
![]()
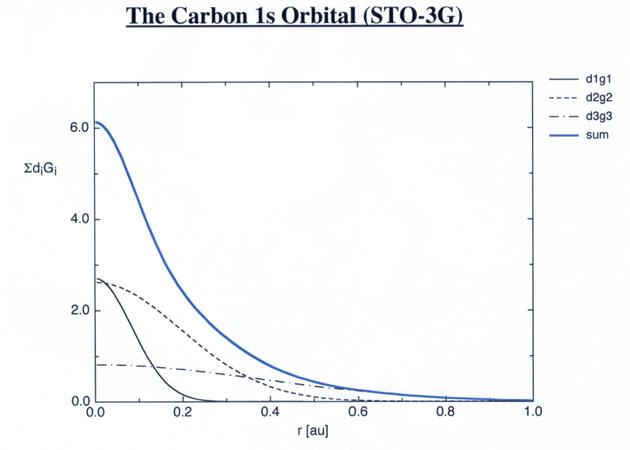
We will use the
STO-3G basis set1 for carbon as an example
(here in a format produced by Gaussian 94):
C 0
S 3
1.00
.7161683735D+02 .1543289673D+00
.1304509632D+02 .5353281423D+00
.3530512160D+01 .4446345422D+00
SP 3
1.00
.2941249355D+01 -.9996722919D-01 .1559162750D+00
.6834830964D+00 .3995128261D+00 .6076837186D+00
.2222899159D+00 .7001154689D+00 .3919573931D+00
The format used is:
Shell_type, No. of primitive Gaussians x, Scale_factor
Orbital exponent ax, Contraction coefficient dx
(repeat x times)
The first line
specifies that the carbon 1s orbital will be described as a sum of three
Gaussian primitives with different exponents a1s,x and coefficients d1s,x. The following
three lines contain the exponent a1s,x and coefficient d1s,x for x=1, 2, and
3. Written in an explicit manner, the 1s basis function for carbon then equates
to:
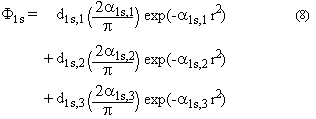
The basis sets for
the 2s and 2p orbitals are also composed of a contraction of three Gaussian
primitives and are specified together in one single "SP"-block. For
the sake of computational efficiency, the 2s and 2p orbitals share the same exponents a2,x , but have different contraction
coefficients. Each line of the basis set description therefore contains only one
exponent (a2s,x = a2p,x ), but two different
coefficients d2s,x and d2p,x. The nodal
characteristics of the 2s-orbital are simulated using one coefficient with
negative sign (-0.09996722919) and two coefficients with positive
(+0.3995128261 and +0.7001154689) sign. Orbital exponents and coefficients have
been optimized first for atomic calculations minimizing the energy of single
atoms. Additional scale factors have, however, then been developed for use of
these atomic basis functions in studies of small organic molecules. The STO-3G
basis set thus contains an appreciable amount of "semiempirical"
character!
How many basis functions do we need for a small organic molecule such as methanol (CH4O)?
%Kjob L301 #P HF/sto-3g GFInput GFPrint methanol basis set 0,1 C1 O2 1 r2 H3 1 r3 2 a3 H4 1 r4 2 a4 3 d4 r2=1.20 r3=1.0 r4=1.0 a3=120. a4=120. d4=180. |
Kjob command kills the job after checking the input The GFInput (“Gaussian Function Input”) output generation keyword causes the current basis set to be printed in a form suitable for use as general basis set input, and can thus be used in adding to or modifying standard basis sets. GFPrint command: This output generation keyword prints the current basis set in tabular form. |
For each of the atomic 1s, 2s and 2p-orbitals,
the STO-3G basis set uses a contraction of three Gaussians. We thus have 5
basis functions for each carbon and oxygen atom, and only one basis function
for each hydrogen, yielding a total of 14 basis functions (and 14
MO-coefficients to vary in the SCF calculations). Each of these basis functions
consists of three Gaussian primitives, and the number of primitives is
therefore 3*14=42. In the output file of Gaussian
98 the number of basis functions and primitives is given by link 301
together with the number of electrons and the core-core repulsion energy:
14 basis functions 42 primitive gaussians
9 alpha electrons 9 beta electrons
nuclear repulsion
energy 40.7431450799 Hartrees.
How "good" is the STO-3G
basis set? The answer to this question depends on the chemical problem at hand.
Structural features of ground state molecules are reproduced quite often with
surprisingly good accuracy at the HF/STO-3G level of theory. Table 1 lists
structural parameters of methanol as predicted by a variety of theoretical
methods.
Table 1. Structural
features of methanol (Cs) as predicted by
several theoretical methods.
Method C-O [Å] C-O-H[˚] bfa
HF/STO-3G 1.432999 103.856 14
HF/3-21G 1.440922 110.336 26
HF/3-21+G 1.454241 113.291 34
HF/6-31G 1.430503 113.410 26
HF/D95 1.437049 113.874 28
HF/6-31G(d) 1.399562 109.448 38
HF/6-31G(d,p) 1.398520 109.650 50
HF/6-31++G(d,p) 1.401386 110.542 62
HF/6-311G(d,p) 1.399240 109.355 60
HF/6-311++G(d,p) 1.400243 110.017 72
HF/6-311G(2df,2pd) 1.396113 109.836 116
HF/cc-pVDZ 1.397818 109.116 48
HF/cc-pVTZ 1.398133 109.937 116
MP2(fc)/cc-pVDZ 1.417284 106.283 48
MP2(fc)/cc-pVTZ 1.418481 107.431 116
exp. 1.421 108.0
a number of basis
functions